Soft x-ray tomography can image an entire cell in its natural hydration state. Determining the location of specific molecular interactions requires that the molecular are labeled with a marker that is visible either in the soft x-ray microscope or in another modality and the data correlated. The latter is the preferred option since direct visualization of a protein using soft x-rays requires it to be labeled with a heavy metal tag (a potentially damaging process to the integrity of the cell structure and molecular organization). Consequently, we have focused our technology development on correlated microscopy, in particular correlated cryogenic confocal microscopy.
Correlated Light and Electron Microscopy (CLEM) is a well-established technique for combining imaging data to generate a composite view of the specimen (Ellisman et al. 2012). After more than 40 years of effort, CLEM is a highly evolved technique and commonplace in the literature. In CLEM, fluorescence microscopy data are used to locate specific features within the specimen, with EM/tomography providing detailed information on the cellular ultrastructure (Martone et al. 2000).
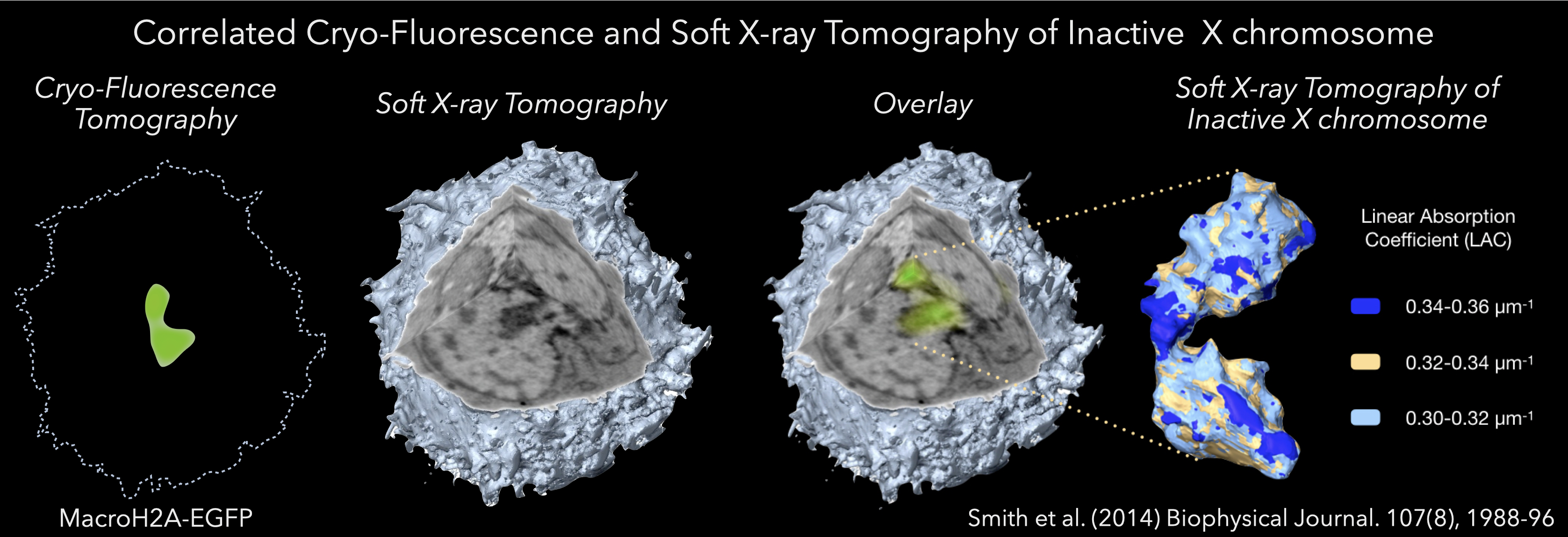
Applying the concept of CLEM was the next logical step after the successful development of SXT. Of course, given SXT generates images of cryopreserved specimens, the fluorescence imaging component must also be carried out at low temperatures (Smith et al., 2013, 2014, 2012). If fluorescence microscopy were carried out at room temperature, and then the cell was frozen, the internal organization would have time to change, perhaps even dramatically, making the combination of data moot (Smith et al. 2013). As a result, cells must be first cryopreserved and then imaged in both the SXM and the fluorescence microscope. Naturally, this requires the cell is to be maintained at low temperature throughout (McDermott et al. 2012a,b). Le Gros et al. addressed this need by developing a new, low-temperature microscope, more details of this work can be found here (Le Gros et al. 2009). In the interest of completeness, we will now give a very brief overview of this development.
The cryogenic light microscope images specimens at liquid nitrogen temperature, using liquid propane (refractive index, RI = 1.32) or iso-pentane (RI = 1.35) as an immersion fluid (Le Gros et al. 2009). Using RI-matched immersion fluid increases light collection efficiency by lowering the amount of light reflected at the interface between different RI (Le Gros et al. 2009). Collecting fluorescence data at cryogenic temperatures has several inherent advantages over room temperature. Firstly, the fluorescence emission spectra of many fluorophores are narrower at low temperature compared with room temperature (Smith et al. 2014a,b). Secondly, and perhaps more importantly, the average working lifetime of fluorescent proteins is extended by a factor of 30 or more at cryogenic temperatures (Moerner and Orrit, 1999). Longer working lifetime allows acquisition of many more images before the fluorescence becomes photo-bleached, and the signal is lost (Smith et al. 2013). This opportunity to collect more images opened up the possibility of adopting a tomographic approach to data acquisition, with the specimen being rotated around a central axis (Cinquin et al. 2014, Smith et al. 2014a,b, Elgass et al. 2015). Reconstructed fluorescence tomography data can achieve isotropic spatial resolution instead of the approximately threefold reduction in resolution that usually occurs along the optical axis (Heintzmann and Cremer, 2002). Tomographic fluorescence imaging, therefore, produces a 3D reconstruction of the fluorescence signal (McDermott et al. 2012a,b, Smith et al. 2014a,b). Since the high numerical aperture optics results in a shallowing of the depth-of-field, at each angular increment, a number of images were collected at varying focal lengths and deconvolved. The result was a deconvolution/tomographic data set that sampled information along the optical axis very well. The only challenge that remained was how to overlay the fluorescence and soft X-ray volumetric data accurately. Fortunately, fluorescence polystyrene beads are visible in both types of data. Simply by adding such beads to the specimen mount the two data types can be aligned independently, and coaligned with great accuracy and precision (Hagen et al. 2012, Smith et al. 2013, Cinquin et al. 2014, Duke et al. 2014, Hagen et al. 2014, Smith et al. 2014a,b, Elgass et al. 2015). As a result, correlated imaging using soft X-rays is the optimal way to locate the position of molecules within the context of a high-resolution 3D reconstruction of a cell and the cellular mesoscale.